· Fabian Schreuder · Data Science Projects · 4 min read
From Tables to Triples: Constructing a Knowledge Graph for Chronic Kidney Disease Data
Leveraging Python, RDFLib, and standard medical ontologies (SNOMED-CT, LOINC) to transform tabular Chronic Kidney Disease data into a structured Knowledge Graph, highlighting data integration and problem-solving.
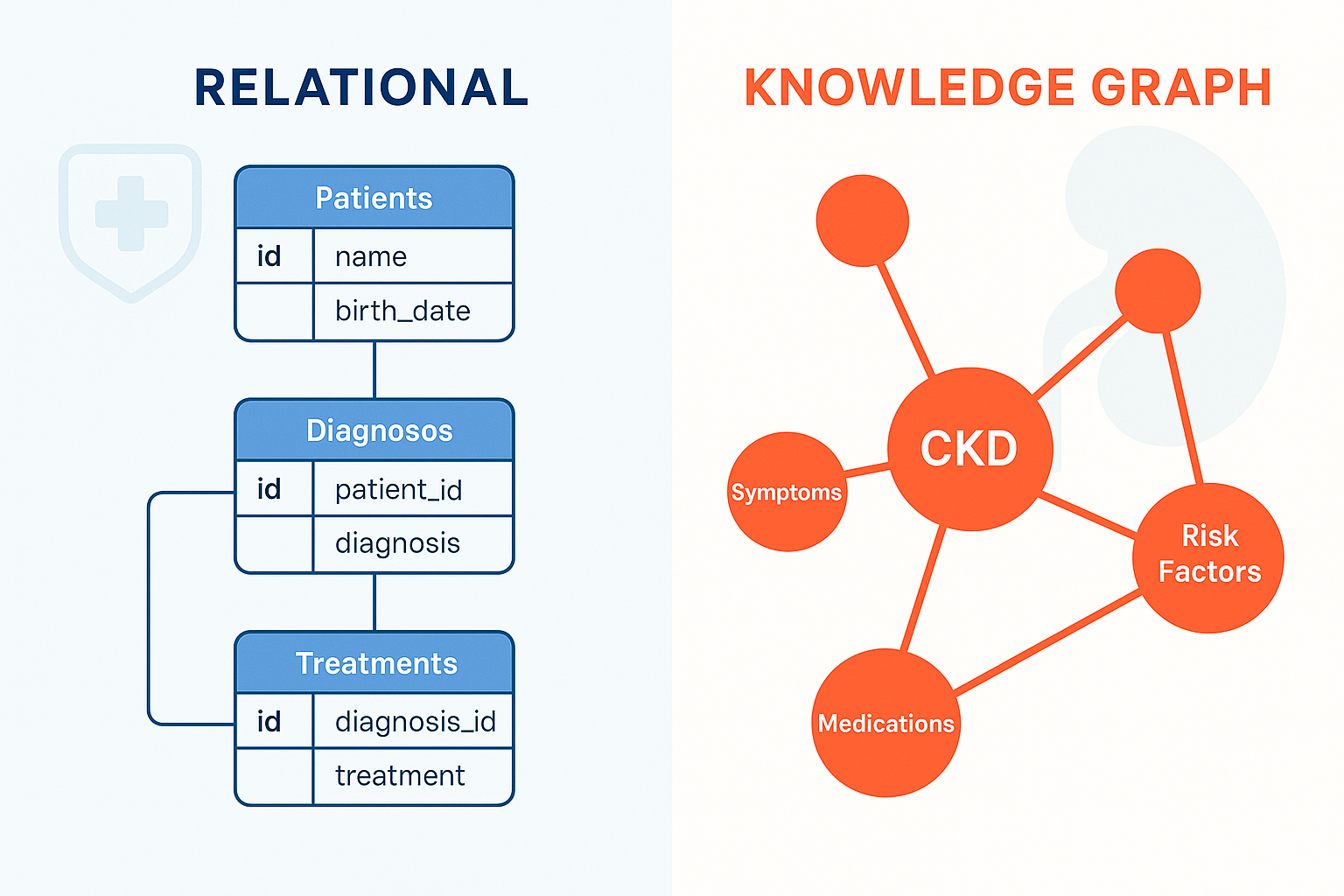
Healthcare data is often complex and highly interconnected. While traditional relational databases excel at storing structured information, they can sometimes struggle to represent the rich, semantic relationships inherent in medical domains like Chronic Kidney Disease (CKD). In a recent project, I explored an alternative approach: building a Knowledge Graph (KG) to model CKD data.
This post outlines the process, challenges, and technical solutions involved in transforming standard tabular data into a more flexible and semantically rich RDF graph structure.
The Limits of Relational Models for Complex Data
Relational databases, with their entity-relationship models, rely on normalized tables, primary keys, and foreign keys. This structure enforces data consistency and is highly efficient for transactional queries. However, representing and querying complex relationships – like correlating different types of clinical observations, lab results, and patient demographics – can become cumbersome, often requiring complex JOIN operations.
For instance, modeling entities like Patient, Measurement, BloodSamples, UrineSamples, and ClinicalObservations in a relational schema defines clear ownership but makes flexible exploration of indirect connections less intuitive.
Embracing Flexibility: The Knowledge Graph Approach
Knowledge Graphs offered a compelling alternative. Using nodes (representing entities like patients or specific measurements) and semantically labeled edges (representing relationships like HAS_MEASUREMENT or CORRELATES_WITH), KGs provide a more natural way to model interconnected data.
The graph structure natively supports querying for relationships and patterns, making it well-suited for exploring the multifaceted nature of conditions like CKD. This flexibility allows for easier integration of diverse data types and discovery of non-obvious connections.
Speaking the Same Language: Leveraging Medical Ontologies
A key step in building a meaningful KG is grounding it in standardized vocabularies, or ontologies. This ensures interoperability and leverages existing domain knowledge. For this CKD project, we carefully selected and mapped our data to established medical ontologies:
- SNOMED-CT: Used for clinical findings and patient attributes (e.g., mapping ‘age’ to SNOMED concept
397669002, and aligning various clinical observations and lab procedures). SNOMED-CT was chosen for its comprehensive coverage of clinical terms and widespread adoption. - LOINC (Logical Observation Identifiers Names and Codes): Used for specific laboratory measurements (e.g., mapping blood pressure to
8462-4and blood glucose to2339-0). LOINC is the standard for identifying lab tests. - QUDT (Quantities, Units, Dimensions and Types): Used for standardizing measurement values and units, ensuring consistency in quantitative data representation.
This ontology mapping process was iterative, requiring careful consideration of domain appropriateness and the adoption level of each ontology.
From Rows to Relationships: The Python Implementation
To perform the conversion from tabular data (likely managed in CSV or similar formats) to the RDF (Resource Description Framework) format required for the KG, I utilized Python, specifically leveraging the powerful Pandas library for data manipulation and RDFLib for creating RDF triples.
The core task involved writing a script that:
- Read the tabular source data.
- Iterated through rows and columns.
- Mapped data points to the corresponding ontology terms (URIs).
- Generated RDF triples (Subject-Predicate-Object) representing the data and its relationships.
- Serialized the resulting graph into an RDF format (e.g., Turtle, XML/RDF).
This process highlighted the practical challenges of handling diverse data types and the crucial importance of understanding the nuances of medical terminologies.
Bridging the Gaps: Handling Custom Properties
Despite the richness of standard ontologies, we encountered specific data points or relationships unique to our dataset that didn’t have direct mappings. Mapping all tabular data proved challenging.
To address this, we adopted a pragmatic solution: creating a custom namespace (which we called MED). Within this namespace, we defined specific properties like med:hasValue to store literal measurement values and med:hasUnit for their corresponding units, when they fell outside the scope of QUDT or other standard representations we could readily implement. This custom namespace acted as a bridge, linking our specific data instances to the broader structure defined by the standard ontologies. This demonstrates a common real-world scenario where perfect mapping isn’t always feasible, requiring practical workarounds.
Key Learnings and My Contribution
This project successfully demonstrated the feasibility of converting tabular CKD data into a structured Knowledge Graph. The resulting KG offers a more flexible and semantically enriched representation compared to a traditional relational model, potentially enabling more sophisticated querying and data analysis in the future.
My specific contributions focused on:
- Researching, identifying, and selecting the appropriate medical ontologies (SNOMED-CT, LOINC, QUDT) for mapping the CKD data elements.
- Developing the Python script using Pandas and RDFLib that performed the core transformation logic, converting the tabular data into RDF triples based on the defined ontology mappings and custom properties.
This project was a valuable exercise in applying semantic web technologies, data modeling principles, and Python programming to a real-world healthcare data challenge, emphasizing the importance of both technical implementation and domain-specific knowledge integration.